
✔ Web-based interface
✔ NGS read library pre-processing and data format conversion
✔ Small RNA knowledge base search
✔ Short-read mapping to genomic reference
✔ De novo sRNA categorization
✔ Argonaute-sRNA affinity inference
✔ Transcriptional and post-transcriptional target prediction
✔ Differential expression analysis
✔ Gene ontology analysis
✔ Network analysis
✔ In silico simulation of sRNA networks
✔ Web-based interface
✔ Machine learning models to predict kinship between small RNA and argonaute proteins from sequence alone
✔ Inference system for small RNA mode of action from sequence alone
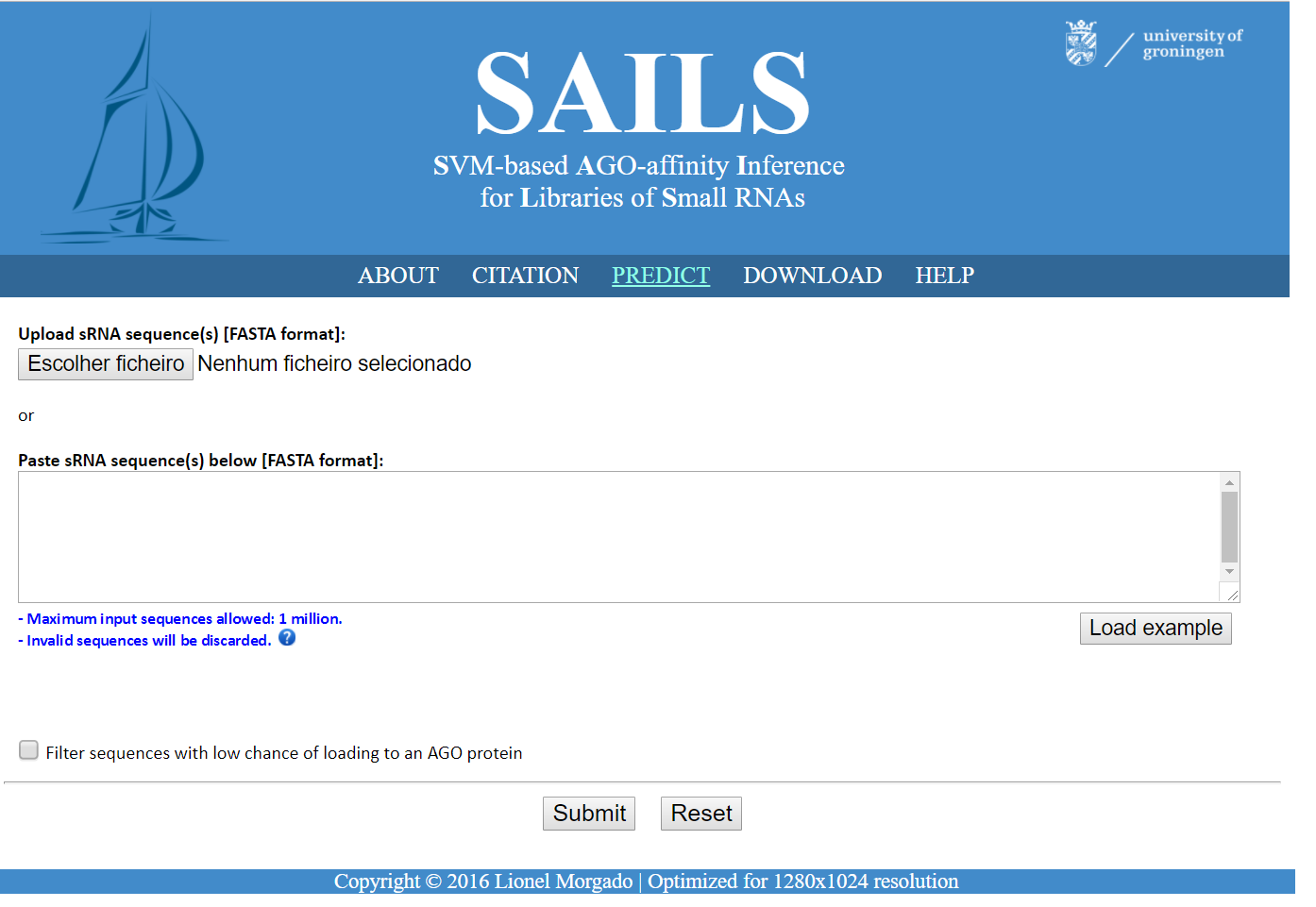
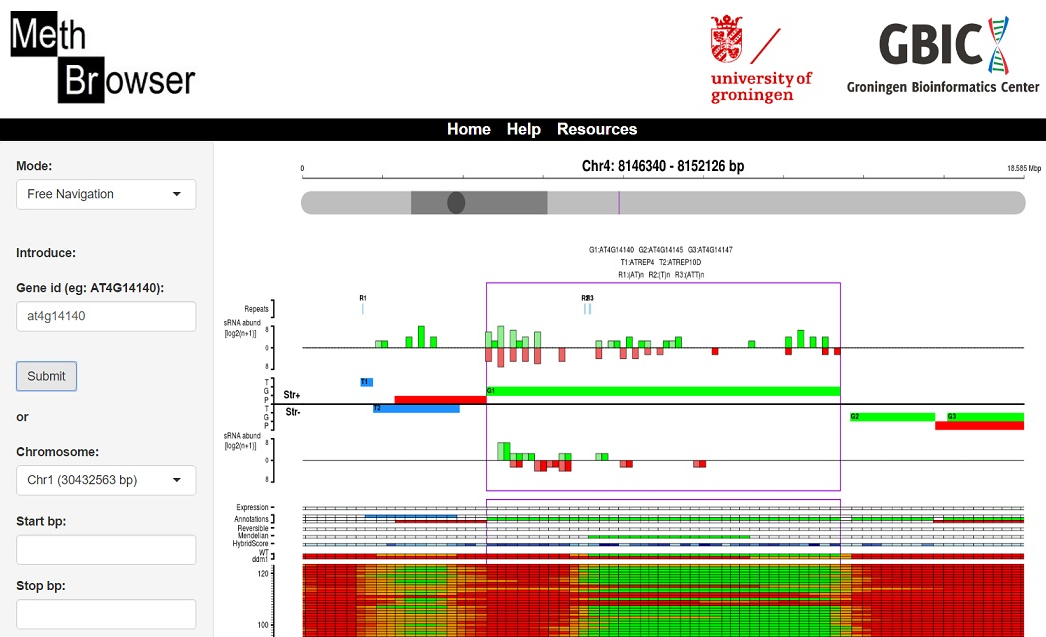
✔ Web-based interface
✔ Visualization of methylomes in populations
✔ Visualization of small RNA abundance
✔ Visualization of genome annotations
✔ Genome-wide querying by genomic coordinates or gene ID
✔ Functionalities to download data in tabular format
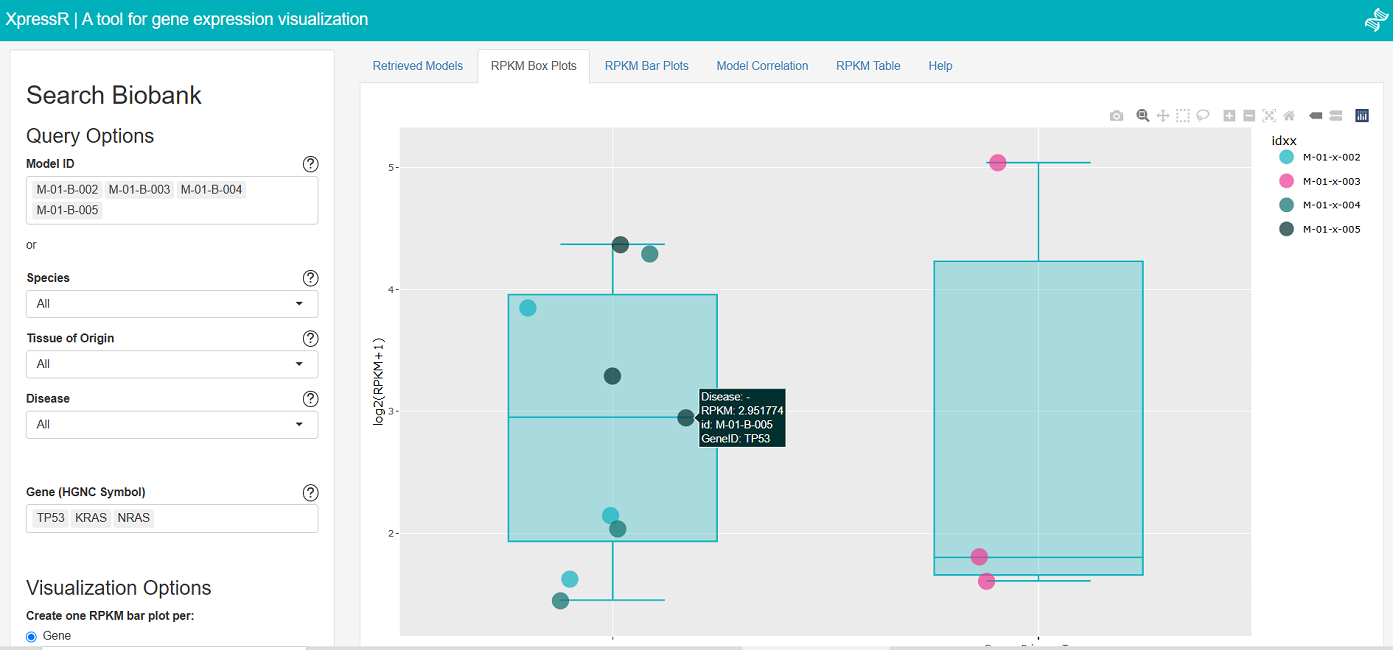
✔ Web-based interface
✔ Querying by model metadata and genes of interest
✔ Output metadata for models retrieved
✔ Interactive boxplots with RPKM values per model, coloured by group
✔ Barplots with RPKM values per gene or per model
✔ Correlation analysis for expression among selected models
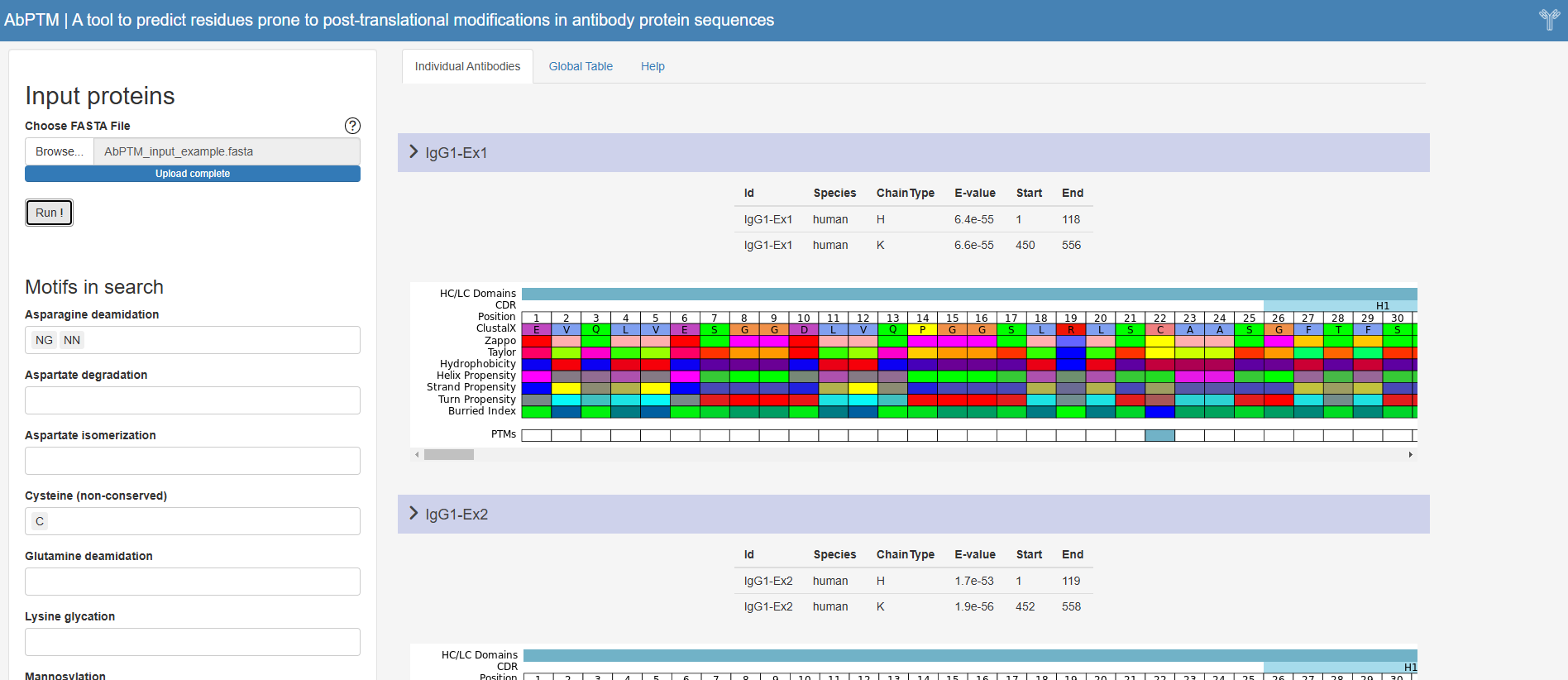
✔ Web-based interface
✔ Numbering and antigen receptor classification
✔ Canonical loop structure annotation (including the detection of the heavy chain, light chain and CDRs)
✔ Detection of PTM prone residues via an embedded motif search
✔ Indication of residue physicochemical properties using multiple popular schemes